elephant.statistics.lvr¶
-
elephant.statistics.lvr(time_intervals, R=array(5.0) * ms, with_nan=False)[source]¶ Calculate the measure of revised local variation LvR for a sequence of time intervals between events.
Given a vector
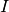 containing a sequence of intervals, the LvR is
defined as:
containing a sequence of intervals, the LvR is
defined as: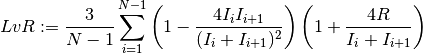
The LvR is a revised version of the Lv, with enhanced invariance to firing rate fluctuations by introducing a refractoriness constant R. The LvR with R=5ms was shown to outperform other ISI variability measures in spike trains with firing rate fluctuatins and sensory stimuli [1].
Parameters: - time_intervalspq.Quantity or np.ndarray or list
Vector of consecutive time intervals.
- Rpq.Quantity or int or float
Refractoriness constant (R >= 0). If no quantity is passed ms are assumed. Default: 5 ms.
- with_nanbool, optional
If True, LvR of a spike train with less than two spikes results in a np.NaN value and a warning is raised. If False, a ValueError exception is raised with a spike train with less than two spikes. Default: True.
Returns: - float
The LvR of the inter-spike interval of the input sequence.
Raises: - ValueError
If an empty list is specified, or if the sequence has less than two entries and with_nan is False.
If a matrix is passed to the function. Only vector inputs are supported.
Warns: - UserWarning
If with_nan is True and the lvr is calculated for a spike train with less than two spikes, generating a np.NaN. If R is passed without any units attached milliseconds are assumed.
References
[1] S. Shinomoto, H. Kim, T. Shimokawa et al. “Relating Neuronal Firing Patterns to Functional Differentiation of Cerebral Cortex” PLOS Computational Biology 5(7): e1000433, 2009.
