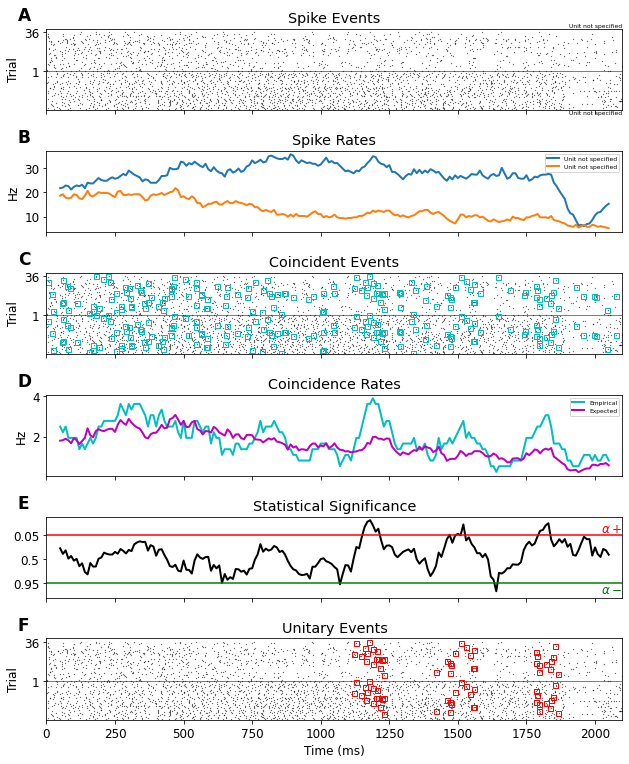
The Unitary Events Analysis¶
The executed version of this tutorial is at https://elephant.readthedocs.io/en/latest/tutorials/unitary_event_analysis.html
The Unitary Events (UE) analysis [1] tool allows us to reliably detect correlated spiking activity that is not explained by the firing rates of the neurons alone. It was designed to detect coordinated spiking activity that occurs significantly more often than predicted by the firing rates of the neurons. The method allows one to analyze correlations not only between pairs of neurons but also between multiple neurons, by considering the various spike patterns across the neurons. In addition, the method allows one to extract the dynamics of correlation between the neurons by perform-ing the analysis in a time-resolved manner. This enables us to relate the occurrence of spike synchrony to behavior.
The algorithm:
- Align trials, decide on width of analysis window.
- Decide on allowed coincidence width.
- Perform a sliding window analysis. In each window:
- Detect and count coincidences.
- Calculate expected number of coincidences.
- Evaluate significance of detected coincidences.
- If significant, the window contains Unitary Events.
- Explore behavioral relevance of UE epochs.
References:
- Grün, S., Diesmann, M., Grammont, F., Riehle, A., & Aertsen, A. (1999). Detecting unitary events without discretization of time. Journal of neuroscience methods, 94(1), 67-79.
[1]:
import random
import string
import numpy as np
import matplotlib.pyplot as plt
import quantities as pq
import neo
import elephant.unitary_event_analysis as ue
# Fix random seed to guarantee fixed output
random.seed(1224)
Next, we download a data file containing spike train data from multiple trials of two neurons.
[2]:
# Download data
!curl https://web.gin.g-node.org/INM-6/elephant-data/raw/master/dataset-1/dataset-1.h5 --output dataset-1.h5 --location
% Total % Received % Xferd Average Speed Time Time Time Current
Dload Upload Total Spent Left Speed
100 369 100 369 0 0 604 0 --:--:-- --:--:-- --:--:-- 605
100 289k 0 289k 0 0 95958 0 --:--:-- 0:00:03 --:--:-- 134k
Load data and extract spiketrains¶
[4]:
block = neo.io.NeoHdf5IO("./dataset-1.h5")
sts1 = block.read_block().segments[0].spiketrains
sts2 = block.read_block().segments[1].spiketrains
spiketrains = np.vstack((sts1,sts2)).T
/home/docs/checkouts/readthedocs.org/user_builds/elephant/conda/v0.10.0/lib/python3.9/site-packages/neo/io/hdf5io.py:63: FutureWarning: NeoHdf5IO will be removed in the next release of Neo. If you still have data in this format, we recommend saving it using NixIO which is also based on HDF5.
warn(warning_msg, FutureWarning)
/home/docs/checkouts/readthedocs.org/user_builds/elephant/conda/v0.10.0/lib/python3.9/site-packages/numpy/core/_asarray.py:171: VisibleDeprecationWarning: Creating an ndarray from ragged nested sequences (which is a list-or-tuple of lists-or-tuples-or ndarrays with different lengths or shapes) is deprecated. If you meant to do this, you must specify 'dtype=object' when creating the ndarray.
return array(a, dtype, copy=False, order=order, subok=True)
Calculate Unitary Events¶
[5]:
UE = ue.jointJ_window_analysis(
spiketrains, bin_size=5*pq.ms, winsize=100*pq.ms, winstep=10*pq.ms, pattern_hash=[3])
plot_ue(spiketrains, UE, significance_level=0.05)
plt.show()
/home/docs/checkouts/readthedocs.org/user_builds/elephant/conda/v0.10.0/lib/python3.9/site-packages/elephant/conversion.py:1168: UserWarning: Binning discarded 1 last spike(s) of the input spiketrain
warnings.warn("Binning discarded {} last spike(s) of the "