Gaussian-Process Factor Analysis (GPFA)¶
Gaussian-process factor analysis (GPFA) is a dimensionality reduction method [1] for neural trajectory visualization of parallel spike trains. GPFA applies factor analysis (FA) to time-binned spike count data to reduce the dimensionality and at the same time smoothes the resulting low-dimensional trajectories by fitting a Gaussian process (GP) model to them.
The input consists of a set of trials (Y), each containing a list of spike trains (N neurons). The output is the projection (X) of the data in a space of pre-chosen dimensionality x_dim < N.
Under the assumption of a linear relation (transform matrix C) between the
latent variable X following a Gaussian process and the spike train data Y with
a bias d and a noise term of zero mean and (co)variance R (i.e.,
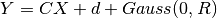 ), the projection corresponds to the
conditional probability E[X|Y].
The parameters (C, d, R) as well as the time scales and variances of the
Gaussian process are estimated from the data using an expectation-maximization
(EM) algorithm.
), the projection corresponds to the
conditional probability E[X|Y].
The parameters (C, d, R) as well as the time scales and variances of the
Gaussian process are estimated from the data using an expectation-maximization
(EM) algorithm.
Internally, the analysis consists of the following steps:
0) bin the spike train data to get a sequence of N dimensional vectors of spike counts in respective time bins, and choose the reduced dimensionality x_dim
1) expectation-maximization for fitting of the parameters C, d, R and the time-scales and variances of the Gaussian process, using all the trials provided as input (c.f., gpfa_core.em())
2) projection of single trials in the low dimensional space (c.f., gpfa_core.exact_inference_with_ll())
3) orthonormalization of the matrix C and the corresponding subspace, for visualization purposes: (c.f., gpfa_core.orthonormalize())
References¶
The code was ported from the MATLAB code based on Byron Yu’s implementation. The original MATLAB code is available at Byron Yu’s website: https://users.ece.cmu.edu/~byronyu/software.shtml
| [1] | Yu MB, Cunningham JP, Santhanam G, Ryu SI, Shenoy K V, Sahani M (2009) Gaussian-process factor analysis for low-dimensional single-trial analysis of neural population activity. J Neurophysiol 102:614-635. |
-
class
elephant.gpfa.gpfa.GPFA(bin_size=array(20.) * ms, x_dim=3, min_var_frac=0.01, tau_init=array(100.) * ms, eps_init=0.001, em_tol=1e-08, em_max_iters=500, freq_ll=5, verbose=False)[source]¶ Apply Gaussian process factor analysis (GPFA) to spike train data
There are two principle scenarios of using the GPFA analysis, both of which can be performed in an instance of the GPFA() class.
In the first scenario, only one single dataset is used to fit the model and to extract the neural trajectories. The parameters that describe the transformation are first extracted from the data using the fit() method of the GPFA class. Then the same data is projected into the orthonormal basis using the method transform(). The fit_transform() method can be used to perform these two steps at once.
In the second scenario, a single dataset is split into training and test datasets. Here, the parameters are estimated from the training data. Then the test data is projected into the low-dimensional space previously obtained from the training data. This analysis is performed by executing first the fit() method on the training data, followed by the transform() method on the test dataset.
The GPFA class is compatible to the cross-validation functions of sklearn.model_selection, such that users can perform cross-validation to search for a set of parameters yielding best performance using these functions.
Parameters: - x_dimint, optional
state dimensionality Default: 3
- bin_sizefloat, optional
spike bin width in msec Default: 20.0
- min_var_fracfloat, optional
fraction of overall data variance for each observed dimension to set as the private variance floor. This is used to combat Heywood cases, where ML parameter learning returns one or more zero private variances. Default: 0.01 (See Martin & McDonald, Psychometrika, Dec 1975.)
- em_tolfloat, optional
stopping criterion for EM Default: 1e-8
- em_max_itersint, optional
number of EM iterations to run Default: 500
- tau_initfloat, optional
GP timescale initialization in msec Default: 100
- eps_initfloat, optional
GP noise variance initialization Default: 1e-3
- freq_llint, optional
data likelihood is computed at every freq_ll EM iterations. freq_ll = 1 means that data likelihood is computed at every iteration. Default: 5
- verbosebool, optional
specifies whether to display status messages Default: False
Raises: - ValueError
If bin_size or tau_init is not a pq.Quantity.
Examples
In the following example, we calculate the neural trajectories of 20 independent Poisson spike trains recorded in 50 trials with randomized rates up to 100 Hz.
>>> import numpy as np >>> import quantities as pq >>> from elephant.gpfa import GPFA >>> from elephant.spike_train_generation import homogeneous_poisson_process >>> data = [] >>> for trial in range(50): >>> n_channels = 20 >>> firing_rates = np.random.randint(low=1, high=100, ... size=n_channels) * pq.Hz >>> spike_times = [homogeneous_poisson_process(rate=rate) ... for rate in firing_rates] >>> data.append((trial, spike_times)) ... >>> gpfa = GPFA(bin_size=20*pq.ms, x_dim=8) >>> gpfa.fit(data) >>> results = gpfa.transform(data, returned_data=['xorth', 'xsm']) >>> xorth = results['xorth']; xsm = results['xsm']
or simply
>>> results = GPFA(bin_size=20*pq.ms, x_dim=8).fit_transform(data, ... returned_data=['xorth', 'xsm'])
Attributes: - valid_data_namestuple of str
Names of the data contained in the resultant data structure, used to check the validity of users’ request
- has_spikes_boolnp.ndarray of bool
Indicates if a neuron has any spikes across trials of the training data.
- params_estimateddict
Estimated model parameters. Updated at each run of the fit() method.
- covTypestr
type of GP covariance, either ‘rbf’, ‘tri’, or ‘logexp’. Currently, only ‘rbf’ is supported.
- gamma(1, #latent_vars) np.ndarray
related to GP timescales of latent variables before orthonormalization by
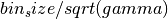
- eps(1, #latent_vars) np.ndarray
GP noise variances
- d(#units, 1) np.ndarray
observation mean
- C(#units, #latent_vars) np.ndarray
loading matrix, representing the mapping between the neuronal data space and the latent variable space
- R(#units, #latent_vars) np.ndarray
observation noise covariance
- fit_infodict
Information of the fitting process. Updated at each run of the fit() method.
- iteration_timelist
containing the runtime for each iteration step in the EM algorithm.
- log_likelihoodslist
log likelihoods after each EM iteration.
- transform_infodict
Information of the transforming process. Updated at each run of the transform() method.
- log_likelihoodfloat
maximized likelihood of the transformed data
- num_binsnd.array
number of bins in each trial
- Corth(#units, #latent_vars) np.ndarray
mapping between the neuronal data space and the orthonormal latent variable space
Methods
fit(self, spiketrains)Fit the model with the given training data. transform(self, spiketrains[, returned_data])Obtain trajectories of neural activity in a low-dimensional latent variable space by inferring the posterior mean of the obtained GPFA model and applying an orthonormalization on the latent variable space. fit_transform(self, spiketrains[, returned_data])Fit the model with spiketrains data and apply the dimensionality reduction on spiketrains. score(self, spiketrains)Returns the log-likelihood of the given data under the fitted model -
fit(self, spiketrains)[source]¶ Fit the model with the given training data.
Parameters: - spiketrainslist of list of neo.SpikeTrain
Spike train data to be fit to latent variables. The outer list corresponds to trials and the inner list corresponds to the neurons recorded in that trial, such that spiketrains[l][n] is the spike train of neuron n in trial l. Note that the number and order of neo.SpikeTrain objects per trial must be fixed such that spiketrains[l][n] and spiketrains[k][n] refer to spike trains of the same neuron for any choices of l, k, and n.
Returns: - selfobject
Returns the instance itself.
Raises: - ValueError
If spiketrains is an empty list.
If spiketrains[0][0] is not a neo.SpikeTrain.
If covariance matrix of input spike data is rank deficient.
-
fit_transform(self, spiketrains, returned_data=['xorth'])[source]¶ Fit the model with spiketrains data and apply the dimensionality reduction on spiketrains.
Parameters: - spiketrainslist of list of neo.SpikeTrain
Refer to the
GPFA.fit()docstring.- returned_datalist of str
Refer to the
GPFA.transform()docstring.
Returns: - np.ndarray or dict
Refer to the
GPFA.transform()docstring.
Raises: - ValueError
Refer to
GPFA.fit()andGPFA.transform().
See also
GPFA.fit- fit the model with spiketrains
GPFA.transform- transform spiketrains into trajectories
-
score(self, spiketrains)[source]¶ Returns the log-likelihood of the given data under the fitted model
Parameters: - spiketrainslist of list of neo.SpikeTrain
Spike train data to be scored. The outer list corresponds to trials and the inner list corresponds to the neurons recorded in that trial, such that spiketrains[l][n] is the spike train of neuron n in trial l. Note that the number and order of neo.SpikeTrain objects per trial must be fixed such that spiketrains[l][n] and spiketrains[k][n] refer to spike trains of the same neuron for any choice of l, k, and n.
Returns: - log_likelihoodfloat
Log-likelihood of the given spiketrains under the fitted model.
-
transform(self, spiketrains, returned_data=['xorth'])[source]¶ Obtain trajectories of neural activity in a low-dimensional latent variable space by inferring the posterior mean of the obtained GPFA model and applying an orthonormalization on the latent variable space.
Parameters: - spiketrainslist of list of neo.SpikeTrain
Spike train data to be transformed to latent variables. The outer list corresponds to trials and the inner list corresponds to the neurons recorded in that trial, such that spiketrains[l][n] is the spike train of neuron n in trial l. Note that the number and order of neo.SpikeTrain objects per trial must be fixed such that spiketrains[l][n] and spiketrains[k][n] refer to spike trains of the same neuron for any choices of l, k, and n.
- returned_datalist of str
The dimensionality reduction transform generates the following resultant data:
‘xorth’: orthonormalized posterior mean of latent variable
‘xsm’: posterior mean of latent variable before orthonormalization
‘Vsm’: posterior covariance between latent variables
‘VsmGP’: posterior covariance over time for each latent variable
‘y’: neural data used to estimate the GPFA model parameters
returned_data specifies the keys by which the data dict is returned.
Default is [‘xorth’].
Returns: - np.ndarray or dict
When the length of returned_data is one, a single np.ndarray, containing the requested data (the first entry in returned_data keys list), is returned. Otherwise, a dict of multiple np.ndarrays with the keys identical to the data names in returned_data is returned.
N-th entry of each np.ndarray is a np.ndarray of the following shape, specific to each data type, containing the corresponding data for the n-th trial:
xorth: (#latent_vars, #bins) np.ndarray
xsm: (#latent_vars, #bins) np.ndarray
y: (#units, #bins) np.ndarray
Vsm: (#latent_vars, #latent_vars, #bins) np.ndarray
VsmGP: (#bins, #bins, #latent_vars) np.ndarray
Note that the num. of bins (#bins) can vary across trials, reflecting the trial durations in the given spiketrains data.
Raises: - ValueError
If the number of neurons in spiketrains is different from that in the training spiketrain data.
If returned_data contains keys different from the ones in self.valid_data_names.
