Spike Pattern Detection and Evaluation (SPADE)¶
SPADE is the combination of a mining technique and multiple statistical tests to detect and assess the statistical significance of repeated occurrences of spike sequences (spatio-temporal patterns, STP).
Given a list of Neo Spiketrain objects, assumed to be recorded in parallel, the SPADE analysis can be applied as demonstrated in this short toy example of 10 artificial spike trains of exhibiting fully synchronous events of order 10.
This modules relies on the implementation of the fp-growth algorithm contained in the file fim.so which can be found here (http://www.borgelt.net/pyfim.html) and should be available in the spade_src folder (elephant/spade_src/). If the fim.so module is not present in the correct location or cannot be imported (only available for linux OS) SPADE will make use of a python implementation of the fast fca algorithm contained in elephant/spade_src/fast_fca.py, which is about 10 times slower.
Examples¶
>>> from elephant.spade import spade
>>> import elephant.spike_train_generation
>>> import quantities as pq
Generate correlated spiketrains.
>>> spiketrains = elephant.spike_train_generation.cpp(
... rate=5*pq.Hz, A=[0]+[0.99]+[0]*9+[0.01], t_stop=10*pq.s)
Mining patterns with SPADE using a binsize of 1 ms and a window length of 1 bin (i.e., detecting only synchronous patterns).
>>> patterns = spade(
... spiketrains=spiketrains, binsize=1*pq.ms, winlen=1, dither=5*pq.ms,
... min_spikes=10, n_surr=10, psr_param=[0,0,3],
... output_format='patterns')['patterns'][0]
>>> import matplotlib.pyplot as plt
>>> for neu in patterns['neurons']:
... label = 'pattern' if neu == 0 else None
... plt.plot(patterns['times'], [neu]*len(patterns['times']), 'ro',
... label=label)
Raster plot of the spiketrains.
>>> for st_idx, spiketrain in enumerate(spiketrains):
... label = 'pattern' if st_idx == 0 else None
... plt.plot(spiketrain.rescale(pq.ms), [st_idx] * len(spiketrain),
... 'k.', label=label)
>>> plt.ylim([-1, len(spiketrains)])
>>> plt.xlabel('time (ms)')
>>> plt.ylabel('neurons ids')
>>> plt.legend()
>>> plt.show()
-
elephant.spade.approximate_stability(concepts, rel_matrix, n_subsets=0, delta=0.0, epsilon=0.0)[source]¶ Approximate the stability of concepts. Uses the algorithm described in Babin, Kuznetsov (2012): Approximating Concept Stability
Parameters: - concepts: list
All the pattern candidates (concepts) found in the spiketrains. Each pattern is represented as a tuple containing (spike IDs, discrete times (window position) of the occurrences of the pattern). The spike IDs are defined as: spike_id=neuron_id*bin_id with neuron_id in [0, len(spiketrains)] and bin_id in [0, winlen].
- rel_matrix: sparse.coo_matrix
A binary matrix with shape (number of windows, winlen*len(spiketrains)). Each row corresponds to a window (order according to their position in time). Each column corresponds to one bin and one neuron and it is 0 if no spikes or 1 if one or more spikes occurred in that bin for that particular neuron. For example, the entry [0,0] of this matrix corresponds to the first bin of the first window position for the first neuron, the entry [0, winlen] to the first bin of the first window position for the second neuron.
- n_subsets: int
Number of subsets of a concept used to approximate its stability. If n_subsets is 0, it is calculated according to to the formula given in Babin, Kuznetsov (2012), proposition 6:
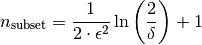
Default: 0
- delta: float, optional
delta: probability with at least
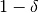 Default: 0.
Default: 0.- epsilon: float, optional
epsilon: absolute error Default: 0.
Returns: - outputlist
List of all the pattern candidates (concepts) given in input, each with the correspondent intensional and extensional stability. Each pattern is represented as a tuple (spike IDs, discrete times of the occurrences of the pattern, intensional stability of the pattern, extensional stability of the pattern). The spike IDs are defined as: spike_id=neuron_id*bin_id with neuron_id in [0, len(spiketrains)] and bin_id in [0, winlen].
Notes
If n_subset is larger than the extent all subsets are directly calculated, otherwise for small extent size an infinite loop can be created while doing the recursion, since the random generation will always contain the same numbers and the algorithm will be stuck searching for other (random) numbers.
-
elephant.spade.concept_output_to_patterns(concepts, winlen, binsize, pv_spec=None, spectrum='#', t_start=array(0.) * ms)[source]¶ Construction of dictionaries containing all the information about a pattern starting from a list of concepts and its associated pvalue_spectrum.
Parameters: - concepts: tuple
Each element of the tuple corresponds to a pattern which it turn is a tuple of (spikes in the pattern, occurrences of the patterns)
- winlen: int
Length (in bins) of the sliding window used for the analysis.
- binsize: pq.Quantity
The time precision used to discretize the spiketrains (binning).
- pv_spec: None or tuple
Contains a tuple of signatures and the corresponding p-value. If equal to None all p-values are set to -1.
- spectrum: {‘#’, ‘3d#’}
- ‘#’: pattern spectrum using the as signature the pair:
(number of spikes, number of occurrences)
- ‘3d#’: pattern spectrum using the as signature the triplets:
(number of spikes, number of occurrence, difference between last and first spike of the pattern)
Default: ‘#’
- t_start: pq.Quantity
t_start of the analyzed spike trains
Returns: - outputlist
List of dictionaries. Each dictionary corresponds to a pattern and has the following entries:
- ‘itemset’:
A list of the spikes in the pattern, expressed in theform of itemset, each spike is encoded by spiketrain_id * winlen + bin_id.
- ‘windows_ids’:
The ids of the windows in which the pattern occurred in discretized time (given byt the binning).
- ‘neurons’:
An array containing the idx of the neurons of the pattern.
- ‘lags’:
An array containing the lags (integers corresponding to the number of bins) between the spikes of the patterns. The first lag is always assumed to be 0 and corresponds to the first spike.
- ‘times’:
An array containing the times (integers corresponding to the bin idx) of the occurrences of the patterns.
- ‘signature’:
A tuple containing two integers (number of spikes of the patterns, number of occurrences of the pattern).
- ‘pvalue’:
The p-value corresponding to the pattern. If n_surr==0, all p-values are set to -1.
-
elephant.spade.concepts_mining(spiketrains, binsize, winlen, min_spikes=2, min_occ=2, max_spikes=None, max_occ=None, min_neu=1, report='a')[source]¶ Find pattern candidates extracting all the concepts of the context, formed by the objects defined as all windows of length winlen*binsize slided along the spiketrains and the attributes as the spikes occurring in each of the window discretized at a time resolution equal to binsize. Hence, the output are all the repeated sequences of spikes with maximal length winlen, which are not trivially explained by the same number of occurrences of a superset of spikes.
Parameters: - spiketrains: list of neo.SpikeTrain
List containing the parallel spike trains to analyze
- binsize: pq.Quantity
The time precision used to discretize the spiketrains (clipping).
- winlen: int
The size (number of bins) of the sliding window used for the analysis. The maximal length of a pattern (delay between first and last spike) is then given by winlen*binsize
- min_spikes: int, optional
Minimum number of spikes of a sequence to be considered a pattern. Default: 2
- min_occ: int, optional
Minimum number of occurrences of a sequence to be considered as a pattern. Default: 2
- max_spikes: int, optional
Maximum number of spikes of a sequence to be considered a pattern. If None no maximal number of spikes is considered. Default: None
- max_occ: int, optional
Maximum number of occurrences of a sequence to be considered as a pattern. If None, no maximal number of occurrences is considered. Default: None
- min_neu: int, optional
Minimum number of neurons in a sequence to considered a pattern. Default: 1
- report: {‘a’, ‘#’, ‘3d#’}, optional
Indicates the output of the function.
- ‘a’:
All the mined patterns
- ‘#’:
Pattern spectrum using as signature the pair: (number of spikes, number of occurrence)
- ‘3d#’:
Pattern spectrum using as signature the triplets: (number of spikes, number of occurrence, difference between the times of the last and the first spike of the pattern)
Default: ‘a’
Returns: - mining_resultsnp.ndarray
- If report == ‘a’:
Numpy array of all the pattern candidates (concepts) found in the spiketrains. Each pattern is represented as a tuple containing (spike IDs, discrete times (window position) of the occurrences of the pattern). The spike IDs are defined as: spike_id=neuron_id*bin_id with neuron_id in [0, len(spiketrains)] and bin_id in [0, winlen].
- If report == ‘#’:
The pattern spectrum is represented as a numpy array of triplets (pattern size, number of occurrences, number of patterns).
- If report == ‘3d#’:
The pattern spectrum is represented as a numpy array of quadruplets (pattern size, number of occurrences, difference between last and first spike of the pattern, number of patterns)
- rel_matrixsparse.coo_matrix
A binary matrix of shape (number of windows, winlen*len(spiketrains)). Each row corresponds to a window (order according to their position in time). Each column corresponds to one bin and one neuron and it is 0 if no spikes or 1 if one or more spikes occurred in that bin for that particular neuron. For example, the entry [0,0] of this matrix corresponds to the first bin of the first window position for the first neuron, the entry [0,winlen] to the first bin of the first window position for the second neuron.
-
elephant.spade.pattern_set_reduction(concepts, ns_signatures, winlen, spectrum, h_subset_filtering=0, k_superset_filtering=0, l_covered_spikes=0, min_spikes=2, min_occ=2)[source]¶ Takes a list concepts and performs pattern set reduction (PSR).
PSR determines which patterns in concepts_psf are statistically significant given any other pattern, on the basis of the pattern size and occurrence count (“support”). Only significant patterns are retained. The significance of a pattern A is evaluated through its signature
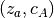 , where
, where 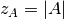 is the size and
is the size and 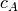 -
the support of A, by either of:
-
the support of A, by either of:- subset filtering: any pattern B is discarded if concepts contains a
superset A of B such that
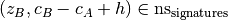
- superset filtering: any pattern A is discarded if concepts contains a
subset B of A such that
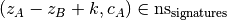
- covered-spikes criterion: for any two patterns A, B with
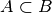 , B is discarded if
, B is discarded if
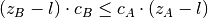 , A is discarded
otherwise;
, A is discarded
otherwise; - combined filtering: combines the three procedures above: takes a list concepts (see output psf function) and performs combined filtering based on the signature (z, c) of each pattern, where z is the pattern size and c the pattern support.
For any two patterns A and B in concepts_psf such that
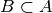 ,
check:
,
check: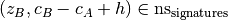 , and
, and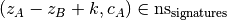 .
.
Then:
- if 1) and not 2): discard B
- if 2) and not 1): discard A
- if 1) and 2): discard B if
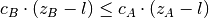 ,
otherwise discard A
,
otherwise discard A
- if neither 1) nor 2): keep both patterns
Assumptions/Approximations:
- a pair of concepts cannot cause one another to be rejected
- if two concepts overlap more than min_occ times, one of them can account for all occurrences of the other one if it passes the filtering
Parameters: - concepts: list
List of concepts, each consisting in its intent and extent
- ns_signatures: list
A list of non-significant pattern signatures (z, c)
- winlen: int
The size (number of bins) of the sliding window used for the analysis. The maximal length of a pattern (delay between first and last spike) is then given by winlen*binsize.
- spectrum: {‘#’, ‘3d#’}
Define the signature of the patterns.
- ‘#’: pattern spectrum using the as signature the pair:
(number of spikes, number of occurrences)
- ‘3d#’: pattern spectrum using the as signature the triplets:
(number of spikes, number of occurrence, difference between last and first spike of the pattern)
- h_subset_filtering: int, optional
Correction parameter for subset filtering Default: 0
- k_superset_filtering: int, optional
Correction parameter for superset filtering Default: 0
- l_covered_spikes: int, optional
Correction parameter for covered-spikes criterion Default: 0
- min_spikes: int, optional
Minimum pattern size Default: 2
- min_occ: int, optional
Minimum number of pattern occurrences Default: 2
Returns: - tuple
A tuple containing the elements of the input argument that are significant according to combined filtering.
- subset filtering: any pattern B is discarded if concepts contains a
superset A of B such that
-
elephant.spade.pvalue_spectrum(spiketrains, binsize, winlen, dither, n_surr, min_spikes=2, min_occ=2, max_spikes=None, max_occ=None, min_neu=1, spectrum='#', surr_method='dither_spikes')[source]¶ Compute the p-value spectrum of pattern signatures extracted from surrogates of parallel spike trains, under the null hypothesis of independent spiking.
- n_surr surrogates are obtained from each spike train by spike dithering
- pattern candidates (concepts) are collected from each surrogate data
- the signatures (number of spikes, number of occurrences) of all patterns are computed, and their occurrence probability estimated by their occurrence frequency (p-value spectrum)
Parameters: - spiketrains: list of neo.SpikeTrain
List containing the parallel spike trains to analyze
- binsize: pq.Quantity
The time precision used to discretize the spiketrains (binning).
- winlen: int
The size (number of bins) of the sliding window used for the analysis. The maximal length of a pattern (delay between first and last spike) is then given by winlen*binsize
- dither: pq.Quantity
Amount of spike time dithering for creating the surrogates for filtering the pattern spectrum. A spike at time t is placed randomly within [t-dither, t+dither] (see also
elephant.spike_train_surrogates.dither_spikes()). Default: 15*pq.s- n_surr: int
Number of surrogates to generate to compute the p-value spectrum. This number should be large (n_surr>=1000 is recommended for 100 spike trains in spiketrains). If n_surr is 0, then the p-value spectrum is not computed. Default: 0
- min_spikes: int, optional
Minimum number of spikes of a sequence to be considered a pattern. Default: 2
- min_occ: int, optional
Minimum number of occurrences of a sequence to be considered as a pattern. Default: 2
- max_spikes: int, optional
Maximum number of spikes of a sequence to be considered a pattern. If None no maximal number of spikes is considered. Default: None
- max_occ: int, optional
Maximum number of occurrences of a sequence to be considered as a pattern. If None, no maximal number of occurrences is considered. Default: None
- min_neu: int, optional
Minimum number of neurons in a sequence to considered a pattern. Default: 1
- spectrum: {‘#’, ‘3d#’}, optional
Defines the signature of the patterns.
- ‘#’: pattern spectrum using the as signature the pair:
(number of spikes, number of occurrence)
- ‘3d#’: pattern spectrum using the as signature the triplets:
(number of spikes, number of occurrence, difference between last and first spike of the pattern)
Default: ‘#’
- surr_method: str
Method that is used to generate the surrogates. You can use every method defined in
elephant.spike_train_surrogates.dither_spikes(). Default: ‘dither_spikes’
Returns: - pv_speclist
- if spectrum == ‘#’:
A list of triplets (z,c,p), where (z,c) is a pattern signature and p is the corresponding p-value (fraction of surrogates containing signatures (z*,c*)>=(z,c)).
- if spectrum == ‘3d#’:
A list of triplets (z,c,l,p), where (z,c,l) is a pattern signature and p is the corresponding p-value (fraction of surrogates containing signatures (z*,c*,l*)>=(z,c,l)).
Signatures whose empirical p-value is 0 are not listed.
-
elephant.spade.spade(spiketrains, binsize, winlen, min_spikes=2, min_occ=2, max_spikes=None, max_occ=None, min_neu=1, approx_stab_pars=None, n_surr=0, dither=array(15.) * ms, spectrum='#', alpha=None, stat_corr='fdr_bh', surr_method='dither_spikes', psr_param=None, output_format='patterns')[source]¶ Perform the SPADE [1-3] analysis for the parallel input spiketrains. They are discretized with a temporal resolution equal to binsize in a sliding window of winlen*binsize.
First, spike patterns are mined from the spiketrains using a technique called frequent itemset mining (FIM) or formal concept analysis (FCA). In this framework, a particular spatio-temporal spike pattern is called a “concept”. It is then possible to compute the stability and the p-value of all pattern candidates. In a final step, concepts are filtered according to a stability threshold and a significance level alpha.
Parameters: - spiketrains: list of neo.SpikeTrain
List containing the parallel spike trains to analyze
- binsize: pq.Quantity
The time precision used to discretize the spiketrains (binning).
- winlen: int
The size (number of bins) of the sliding window used for the analysis. The maximal length of a pattern (delay between first and last spike) is then given by winlen*binsize
- min_spikes: int, optional
Minimum number of spikes of a sequence to be considered a pattern. Default: 2
- min_occ: int, optional
Minimum number of occurrences of a sequence to be considered as a pattern. Default: 2
- max_spikes: int, optional
Maximum number of spikes of a sequence to be considered a pattern. If None no maximal number of spikes is considered. Default: None
- max_occ: int, optional
Maximum number of occurrences of a sequence to be considered as a pattern. If None, no maximal number of occurrences is considered. Default: None
- min_neu: int, optional
Minimum number of neurons in a sequence to considered a pattern. Default: 1
- approx_stab_pars: dict or None, optional
Parameter values for approximate stability computation.
- ‘n_subsets’: int
Number of subsets of a concept used to approximate its stability. If n_subsets is 0, it is calculated according to to the formula given in Babin, Kuznetsov (2012), proposition 6:
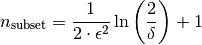
- ‘delta’: float, optional
delta: probability with at least
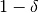
- ‘epsilon’: float, optional
epsilon: absolute error
- ‘stability_thresh’: None or list of float
List containing the stability thresholds used to filter the concepts. If stability_thresh is None, then the concepts are not filtered. Otherwise, only concepts with
intensional stability is greater than stability_thresh[0] or
extensional stability is greater than stability_thresh[1]
are further analyzed.
- n_surr: int, optional
Number of surrogates to generate to compute the p-value spectrum. This number should be large (n_surr >= 1000 is recommended for 100 spike trains in spiketrains). If n_surr == 0, then the p-value spectrum is not computed. Default: 0
- dither: pq.Quantity, optional
Amount of spike time dithering for creating the surrogates for filtering the pattern spectrum. A spike at time t is placed randomly within [t-dither, t+dither] (see also
elephant.spike_train_surrogates.dither_spikes()). Default: 15*pq.ms- spectrum: {‘#’, ‘3d#’}, optional
Define the signature of the patterns.
- ‘#’: pattern spectrum using the as signature the pair:
(number of spikes, number of occurrences)
- ‘3d#’: pattern spectrum using the as signature the triplets:
(number of spikes, number of occurrence, difference between last and first spike of the pattern)
Default: ‘#’
- alpha: float, optional
The significance level of the hypothesis tests performed. If alpha is None, all the concepts are returned. If 0.<alpha<1., the concepts are filtered according to their signature in the p-value spectrum. Default: None
- stat_corr: str
Method used for multiple testing. See:
test_signature_significance()Default: ‘fdr_bh’- surr_method: str
Method to generate surrogates. You can use every method defined in
elephant.spike_train_surrogates.surrogates(). Default: ‘dither_spikes’- psr_param: None or list of int
- This list contains parameters used in the pattern spectrum filtering:
- psr_param[0]: correction parameter for subset filtering
(see h_subset_filtering in
pattern_set_reduction()).- psr_param[1]: correction parameter for superset filtering
(see k_superset_filtering in
pattern_set_reduction()).- psr_param[2]: correction parameter for covered-spikes criterion
(see l_covered_spikes in
pattern_set_reduction()).
- output_format: {‘concepts’, ‘patterns’}
Distinguish the format of the output (see Returns). Default: ‘patterns’
Returns: - outputdict
- ‘patterns’:
If output_format is ‘patterns’, see the return of
concept_output_to_patterns()If output_format is ‘concepts’, then output[‘patterns’] is a tuple of patterns which in turn are tuples of
- spikes in the pattern
- occurrences of the pattern
For details see
concepts_mining().if stability is calculated, there are also:
- intensional stability
- extensional stability
For details see
approximate_stability().- ‘pvalue_spectrum’ (only if n_surr > 0):
- A list of signatures in tuples format:
- size
- number of occurrences
- duration (only if spectrum==’3d#’)
- p-value
- ‘non_sgnf_sgnt’: list
Non significant signatures of ‘pvalue_spectrum’.
Notes
If detected, this function will use MPI to parallelize the analysis.
References
- [1] Torre, E., Picado-Muino, D., Denker, M., Borgelt, C., & Gruen, S.(2013)
- Statistical evaluation of synchronous spike patterns extracted by frequent item set mining. Frontiers in Computational Neuroscience, 7.
- [2] Quaglio, P., Yegenoglu, A., Torre, E., Endres, D. M., & Gruen, S.(2017)
- Detection and Evaluation of Spatio-Temporal Spike Patterns in Massively Parallel Spike Train Data with SPADE. Frontiers in Computational Neuroscience, 11.
- [3] Stella, A., Quaglio, P., Torre, E., & Gruen, S. (2019).
- 3d-SPADE: Significance evaluation of spatio-temporal patterns of various temporal extents. Biosystems, 185, 104022.
Examples
The following example applies SPADE to spiketrains (list of neo.SpikeTrain).
>>> from elephant.spade import spade >>> import quantities as pq >>> binsize = 3 * pq.ms # time resolution to discretize the spiketrains >>> winlen = 10 # maximal pattern length in bins (i.e., sliding window) >>> result_spade = spade(spiketrains, binsize, winlen)
-
elephant.spade.test_signature_significance(pv_spec, concepts, alpha, winlen, corr='fdr_bh', report='spectrum', spectrum='#')[source]¶ Compute the significance spectrum of a pattern spectrum.
Given pvalue_spectrum pv_spec as a list of triplets (z,c,p), where z is pattern size, c is pattern support and p is the p-value of the signature (z,c), this routine assesses the significance of (z,c) using the confidence level alpha.
Bonferroni or FDR statistical corrections can be applied.
Parameters: - pv_spec: list
A list of triplets (z,c,p), where z is pattern size, c is pattern support and p is the p-value of signature (z,c)
- concepts: list of tuple
Output of the concepts mining for the original data.
- alpha: float
Significance level of the statistical test
- winlen: int
Size (number of bins) of the sliding window used for the analysis
- corr: str, optional
Method used for testing and adjustment of pvalues. Can be either the full name or initial letters. Available methods are:
‘bonferroni’ : one-step correction
‘sidak’ : one-step correction
‘holm-sidak’ : step down method using Sidak adjustments
‘holm’ : step-down method using Bonferroni adjustments
‘simes-hochberg’ : step-up method (independent)
‘hommel’ : closed method based on Simes tests (non-negative)
‘fdr_bh’ : Benjamini/Hochberg (non-negative)
‘fdr_by’ : Benjamini/Yekutieli (negative)
‘fdr_tsbh’ : two stage fdr correction (non-negative)
‘fdr_tsbky’ : two stage fdr correction (non-negative)
‘’ or ‘no’: no statistical correction
For further description see: https://www.statsmodels.org/stable/generated/statsmodels.stats.multitest.multipletests.html Default: ‘fdr_bh’
- report: {‘spectrum’, ‘significant’, ‘non_significant’}, optional
Format to be returned for the significance spectrum:
- ‘spectrum’: list of triplets (z,c,b), where b is a boolean specifying
whether signature (z,c) is significant (True) or not (False)
- ‘significant’: list containing only the significant signatures (z,c) of
pvalue_spectrum
‘non_significant’: list containing only the non-significant signatures
- spectrum: {‘#’, ‘3d#’}, optional
Defines the signature of the patterns.
- ‘#’: pattern spectrum using the as signature the pair:
(number of spikes, number of occurrence)
- ‘3d#’: pattern spectrum using the as signature the triplets:
(number of spikes, number of occurrence, difference between last and first spike of the pattern)
Default: ‘#’
Returns: - sig_spectrumlist
Significant signatures of pvalue_spectrum, in the format specified by report
