elephant.spike_train_correlation.correlation_coefficient¶
-
elephant.spike_train_correlation.correlation_coefficient(binned_spiketrain, binary=False, fast=True)[source]¶ Calculate the NxN matrix of pairwise Pearson’s correlation coefficients between all combinations of N binned spike trains.
For each pair of spike trains
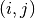 , the correlation coefficient
, the correlation coefficient
![C[i,j]](../../../_images/math/bf98d44b10da7feeb3ab48513f947f6939e208a9.png) is obtained by binning
is obtained by binning 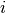 and
and 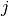 at the
desired bin size. Let
at the
desired bin size. Let 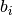 and
and 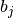 denote the binned spike
trains and
denote the binned spike
trains and 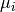 and
and 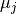 their respective means. Then
their respective means. Then![C[i,j] = <b_i-\mu_i, b_j-\mu_j> /
\sqrt{<b_i-\mu_i, b_i-\mu_i> \cdot <b_j-\mu_j, b_j-\mu_j>}](../../../_images/math/09c96cd3939401b9a6cde741c06d65e88db0698e.png)
where <., .> is the scalar product of two vectors.
For an input of N spike trains, an N x N matrix is returned. Each entry in the matrix is a real number ranging between -1 (perfectly anti-correlated spike trains) and +1 (perfectly correlated spike trains). However, if k-th spike train is empty, k-th row and k-th column of the returned matrix are set to np.nan.
If binary is True, the binned spike trains are clipped to 0 or 1 before computing the correlation coefficients, so that the binned vectors
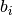 and
and 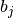 are binary.
are binary.Visualization of this function is covered in Viziphant:
viziphant.spike_train_correlation.plot_corrcoef().Parameters: - binned_spiketrain(N, ) elephant.conversion.BinnedSpikeTrain
A binned spike train containing the spike trains to be evaluated.
- binarybool, optional
If True, two spikes of a particular spike train falling in the same bin are counted as 1, resulting in binary binned vectors
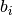 . If
False, the binned vectors
. If
False, the binned vectors 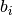 contain the spike counts per bin.
Default: False.
contain the spike counts per bin.
Default: False.- fastbool, optional
If fast=True and the sparsity of binned_spiketrain is > 0.1, use np.corrcoef(). Otherwise, use memory efficient implementation. See Notes[2] Default: True.
Returns: - C(N, N) np.ndarray
The square matrix of correlation coefficients. The element
![C[i,j]=C[j,i]](../../../_images/math/709f60b89db15874f0deced9152a19286b30d59d.png) is the Pearson’s correlation coefficient between
binned_spiketrain[i] and binned_spiketrain[j].
If binned_spiketrain contains only one neo.SpikeTrain, C=1.0.
is the Pearson’s correlation coefficient between
binned_spiketrain[i] and binned_spiketrain[j].
If binned_spiketrain contains only one neo.SpikeTrain, C=1.0.
Raises: - MemoryError
When using fast=True and binned_spiketrain shape is large.
Warns: - UserWarning
If at least one row in binned_spiketrain is empty (has no spikes).
See also
Notes
- The spike trains in the binned structure are assumed to cover the complete time span [t_start, t_stop) of binned_spiketrain.
- Using fast=True might lead to MemoryError. If it’s the case, switch to fast=False.
Examples
Generate two Poisson spike trains
>>> import neo >>> from quantities import s, Hz, ms >>> from elephant.spike_train_generation import homogeneous_poisson_process >>> from elephant.conversion import BinnedSpikeTrain >>> st1 = homogeneous_poisson_process( ... rate=10.0*Hz, t_start=0.0*s, t_stop=10.0*s) >>> st2 = homogeneous_poisson_process( ... rate=10.0*Hz, t_start=0.0*s, t_stop=10.0*s) >>> cc_matrix = correlation_coefficient(BinnedSpikeTrain([st1, st2], ... bin_size=5*ms)) >>> print(cc_matrix[0, 1]) 0.015477320222075359
